
|
BIOL 5130
Evolution
Phil
Ganter
301 Harned Hall
963-5782 |
|
From Humbolt State Forest in California |
Genetic Drift
Lecture
03
Email me
Assignments Page
Back
to:
Genetic
Drift is a consequence of chance events
- random in direction, perhaps better
called "genetic chance"
- Represents a departure from Hardy-Weinberg
assumptions
- We will begin the examination
of the consequences of drift by discussing it in isolation from other
H-W departures but it happens no matter which other departures are occurring
- i. e., selection and drift,
mutation and drift, migration and drift, etc. all occur
- the rate of change due to random
chance is dependent on population size (inversely dependent)
Fixation
- fixation is the loss of all alleles for a locus save one (this allele said
to be fixed)
- drift will produce fixation given
enough time
- which allele will be fixed?
- without selection favoring a
single allele, all alleles have the same probability of being fixed
- in a population there are 2N
alleles (N = population size), so any allele present in the population
today has a probability of being fixed some time in the future equal to
1/2N
- under
what circumstance are small populations most likely?
- rare species are, by definition,
found only in small population sizes (threatened or endangered species
belong in this category also)
- even species characterized by
large population sizes may be subjected to small population size dynamics
when:
- a new population is founded
(called a Founder Event)
- the founders may lack
all of the alleles of the source population or may have a
gene
frequency that differs from the source population
- what is the chance that a founding
population will be homozygous at a given locus?
- assume only two alleles in the source
population, n = number of founders, and the chance of drawing
an individual with allele A is p (a is q) then
Pr(homozygosity) = p2n
+ q2n
- the probability falls quickly
as n, the number of founders, increases but this may still be an important
source of variation between populations if the number of founders does
tend to be small
- environmental extremes cause
populations to crash, even if they subsequently regrow
to their
previous
size
- a crash followed by an increase
in population size is called a Bottleneck
- the amount of time it takes to fix an allele
is also dependent on population size, directly dependent this time
Coalescence
- looking back, instead of ahead,
is follows that all alleles in the population today are the descendents of
a single allele that existed some time in the past
- the alleles in today's population
are said to coalesce to this single ancestor
- ancestral allele is called the
coalescent
- how long ago the coalescent existed
depends, assuming that it did exist, on the population size (now and in the
past), selection strength, mating system, and other aspects of the life history
of the organism being studied
- currently this approach is being
used to estimate the history of one or more populations, something once
impossible to do without explicit historical records (almost never available)
- This link will take you to the
Lamarc
site, the website for one team of researchers who make available software
to estimate population parameters based on the coalescent
- (LAMARC stands for Likelihood
Analysis with Metropolis Algorithm using Random Coalescence - how's that
for creative acronyming, if not totally accurate spelling)
Heterozygosity and Drift
- We have seen that drift can cause fixation, but
what is its effect in larger populations?
- to get the idea of what drift will do to a population
over time, consider what will happen in one generation to the frequency of
homozygotes in the population (compared to the same frequency in the previous
population) in the case of two alleles
- f is the proportion of homozygotes in the current
generation (which is at H-W equilibrium as selection, etc. are not occurring),
so
f = p2 +
q2
- you can generalize this for more than two alleles
by using p as the probability of homozygosity at each of i alleles so that:
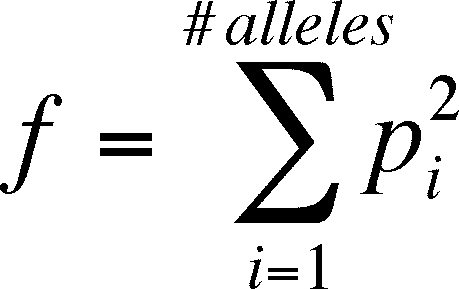
- there
are two ways to become a homozygote in the next generation, if we consider
the
possibility
of self-fertilization:
- two gametes from the same individual combine, which
has a probability of 1/2N (N = population size)
- two gametes with the same gene but from different
individuals combine. The probability of this is the probability of
picking a second gamete that is not from the same individual, which is =
1 - (1/2N) [notice that the two probabilities so far add to one]. If
we are producing homozygotes, we have to multiply this probability
times the probability that the individual is a homozygote, f
- the probability of being a homozygote in the next
generation, f' is then the sum of these two independent probabilities of
producing a homozygote
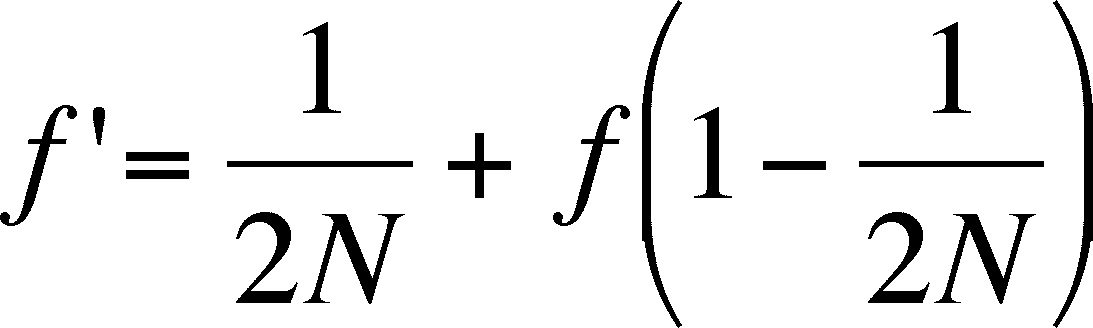
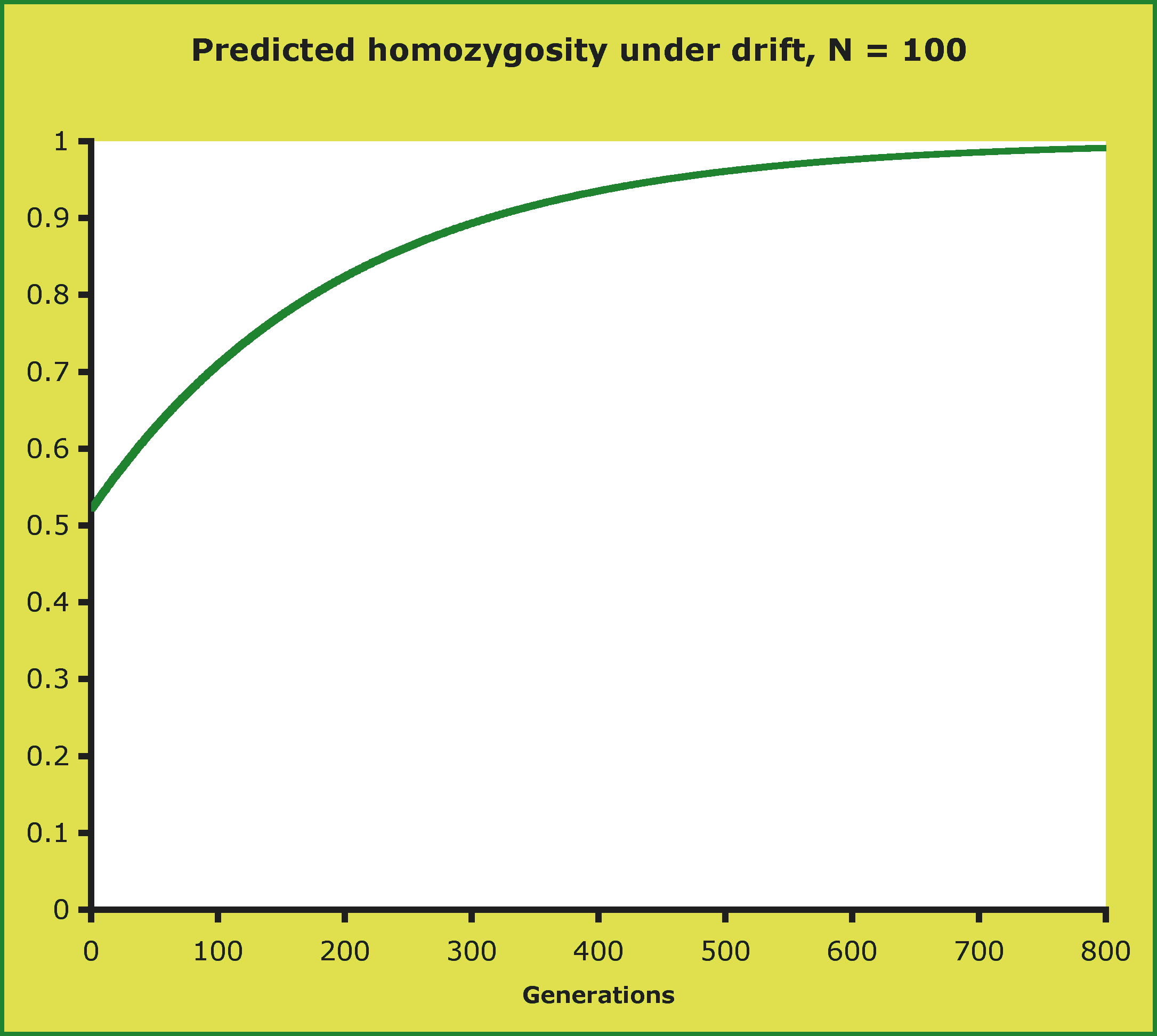
- if you begin with p = .6 and N = 100, homozygosity will drift upward, reaching
near fixation in hundreds of generations:
Heterozygosity
- measure
of genetic variation
- for any locus, it is the chance that two alleles,
drawn at random, are different
- logically, there is a complementary relationship
between H and f, such that H = 1 - f and
(substituting in the equation above)
- in the case of more than two alleles (modified
from general case for f above):
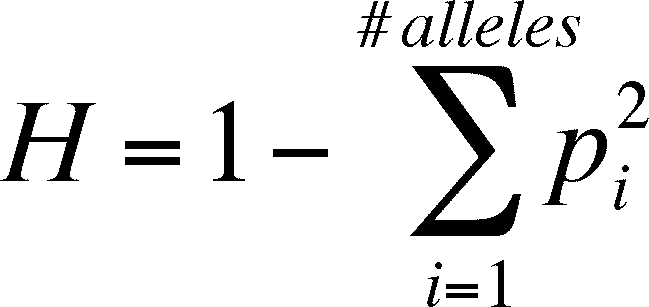
- the change in heterozygosity due to drift
decreases over time and is predicted by:
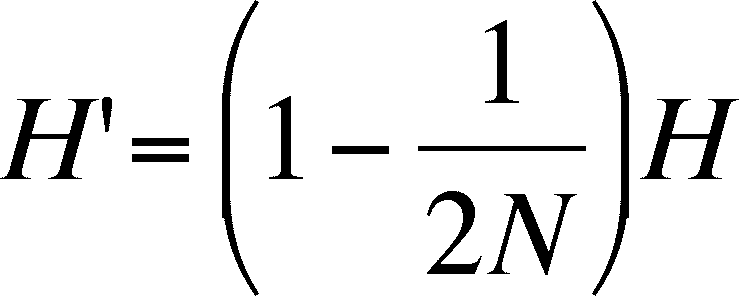
- really confusing terminology due to meaning
of heterozygous individuals (populations can be
heterozygous
even
if all
the
individual within are homozygous), better term is gene diversity
- sequence data has meant that we can look at genetic
variation by site and this variation is usually measured as pi,
the total number of nucleotide differences between two sequences
divided by the length of the sequence (to facilitate comparisons
among sequences
of
different sizes
Mutation and Drift
Some aspects of point mutations (also
called substitutions)
Relative Frequency (estimated from
changes in pseudogenes)
|
Before
Mutation |
|
A |
T |
C |
G |
|
- |
1.3 |
2.0 |
6.3 |
T |
1.4 |
- |
6.4 |
2.2 |
C |
1.4 |
2.5 |
- |
1.6 |
G |
2.8 |
1.0 |
1.3 |
- |
Transitions |
Transversions |
As you can see, transitions are more common
than transversions (and not symmetric!)
redundancy in the genetic code
means that many third position substitutions are selectively neutral as
they cannot affect the primary sequence of the encoded protein
Insertions and Deletions (Indels)
-
Analysis of these are often
problematic and the most common response is to ignore them when comparing
sequences (is this acceptable beyond its practicality?)
-
can't tell if an indel
of more than one bp is a single evolutionary event or multiple events
-
if the location of indels
is not random, they will tend to overlap and make sorting the events
that produce the observed pattern difficult or impossbile
-
when doing alignments of two
or more sequences, it is usually to create multiple indels so that the
base pairs aligned are homologous base pairs
-
however, if you insert enough
indels, you can match most sequences without any substitutions
-
so when to insert an indel
and when to accept that a substitution has occurred is often determined
by penalizing an indel more (in terms of evolutionary distance) than
a substitution (which is usually a distance of 1 or close to it)
-
when an indel lowers the
overall score for the entire sequence comparison, it is accepted, when
it does not it is rejected
Mutations at the
gene level (and at the position level in a sequence) are often subject to
reversion
-
this is not expected for complex
characters governed by many loci and many alleles
-
the next mutation to the
entire system of genes contributing to the phenotype is unlikely to
be at the same position as the initial mutation and so is not likely
to cause reversion
-
as more mutation accrue
to the entire system, the likelihood that a mutation that cause reversion
becomes even less likely
-
for characters determined by
one or a few genes, reversion becomes more likely
-
ex. - ability of a microbe
to utilize a particular carbon compound as a source of energy
-
often this is governed
by only a few genes (a transport gene in the membrane, an enzyme
to
split polymers into monomers, and perhaps an enzyme to modify the
monomer into a substrate found in glycolysis or the Krebs cycle)
-
so given that p is frequency
of A, m is the rate of change from A to a, and a is the frequency of change
from a to A is n
-
m and n are usually their
Greek equivalents but these are not always displayed well in all browsers,
so I will use English here
-
m and n are rates but are
used here as the proportion of each allele that will mutate into the
other allele during a single time interval (size dependent on the
situation)
pt+1
= pt (1 - m) + (1 - pt)n
p* = p*(1 - m) + n(1
- p*)
= p* - mp* + n - np*
p* - p* + mp* + np* =
n
p*(m + n) = n
p* = n/(m + n)
-
so, as m increases, p* decreases
and as n increases, p* also increases, although not in a linear fashion
-
Drift and Mutation can balance
on another, such that fixation is staved off by the appearance of the
allele
through mutation. The equilibruim values for homozygosity and
heterozygosity, where disappearance is balanced by mutation, depends
on the mutation rate
and the population size:
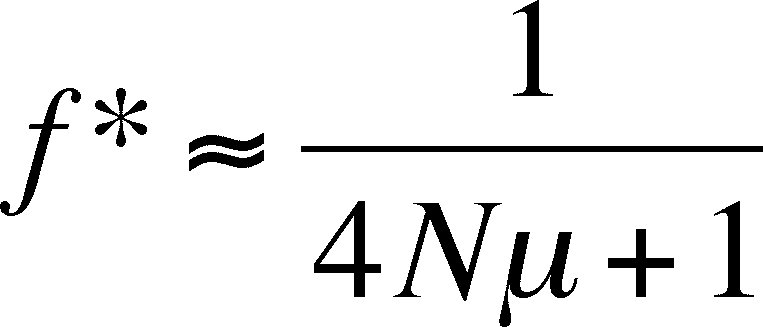
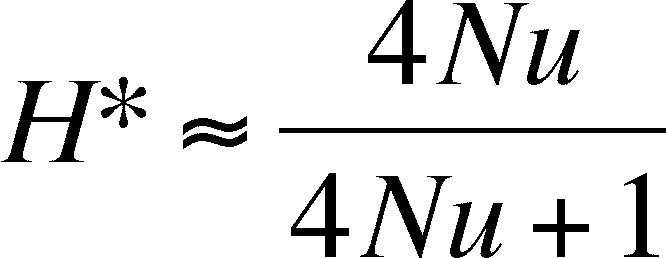
Effective Population Size
-
population size is critical when assessing the effects of drift,
so it is meaningful to ask how to measure it
-
Effective Population Size (Ne)
-
the estimation of the probability that two randomly chosen
allels are the same is affected by:
-
sex ratio -
each sex contributes half of the alleles to the next generation, even
when one
sex is rare in the present generation. The
rare sex's genotypic ratios will then have a greater influence than
the more numerous sex.
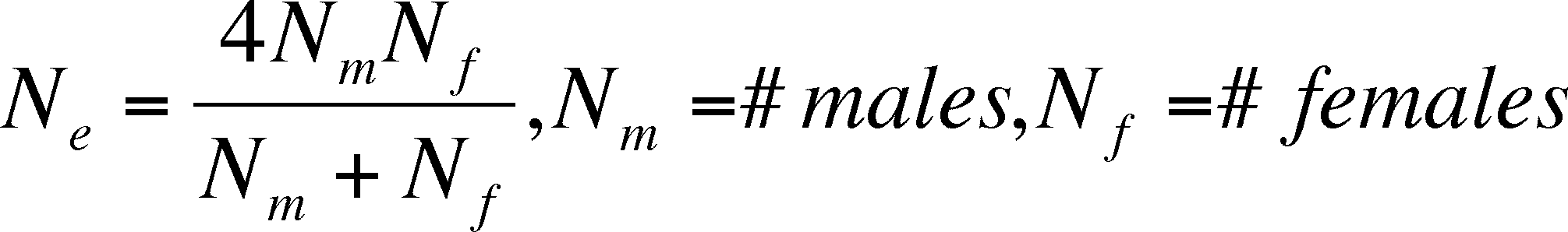
-
you can link to an old worksheet on effective population size
done for the ecology class several years ago by clicking on this link (use
your browser's back button to get back to here)
-
population sub-division and mating groups -
both of these will effectively lower the population Ne but
the corrections needed are too complex to discuss here
-
variation in fecundity - when some females have more offspring
than others, those with high fertitility have a greater effect on the genotypic
composition of the next generation than those with lower fertility
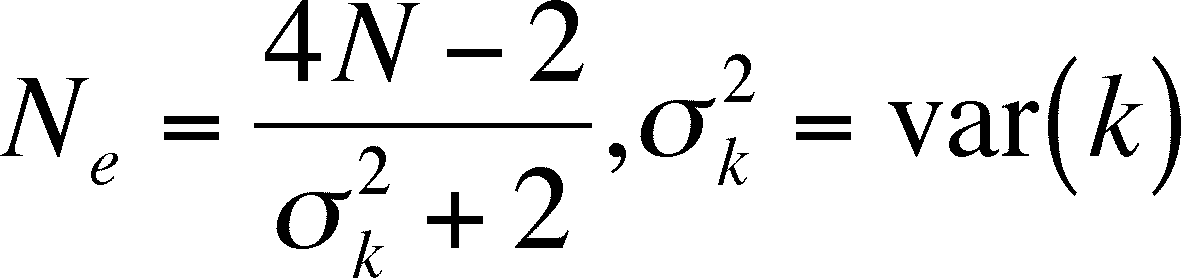
Natural Selection and Drift
-
Drift will dominate when there is
no selective pressure on an allele (or very small selective advantage or
disadvantage)
-
Adaptation is produces by natural
selection, but much evolution might be due to drift
-
e. g., change in non-coding
sequences (that also have no other function) such as intergenic spacers,
pseudogenes, and some third positions in codons
Neutral Theory covers evolution
without selection
-
argued that genetic load, if multiplicative,
of even small selection coefficients, within a large gene would make one
allele almost lethal in most cases
-
molecular clock is dependent on
neutral drift
-
rates have been shown to be constant
for some genes but not the same rate for all genes evolving at a constant
rate.
-
in some cases, loci have been shown
not to be very "clock-like" in that they have not always evolved at a constant
rate (environmental genes)
-
neutral theory and the clock
-
Constraints
-
a question under investigation
now and for some time has been "what constrains evolution?"
-
functional constraints - a protein
must do its job and changes that interfere with that are not likely
to have high fitness.
-
developmental constraint - a
change to a locus must still interact with other loci to produce a viable
phenotype
-
this second constraint has been
of growing interest as we begin to understand development (to some degree
and only in some model organisms)
-
remains at this time an area of
potential constraint more than one of demonstrated constraints.
Studying natural selection
without measuring fitness
-
this is an area confined, at this
point, to molecular data
-
remember, selection does not always
have the same effect
-
stabilizing selection (sometimes
called negative selection in that it resists change) will reduce the number
of changes made over neutral drift
-
directional selection (sometimes
called positive selection in that it promotes changes to the molecule)
will increase number of changes over neutral drift
-
dN/dS ratio (Non-synonymous / Synonymous)
a measure of change within proteins potentially subject to natural selection
(Non-synonymous) to Synonymous changes not subject to selection (= neutral
change)
-
Convergence
-
when similar adaptations arise independently
in different lineages, changes that appear in all lineages are convergent
evolution and can be interpreted and evidence of stabilizing selection
at that site.
-
Codon Bias
Last updated January
28, 2008
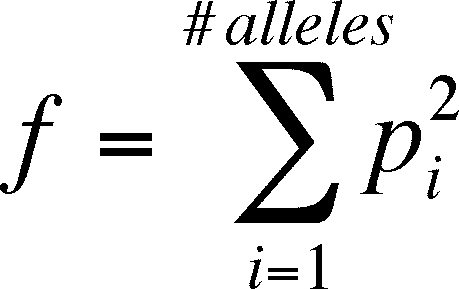
![]()

![]()
![]()
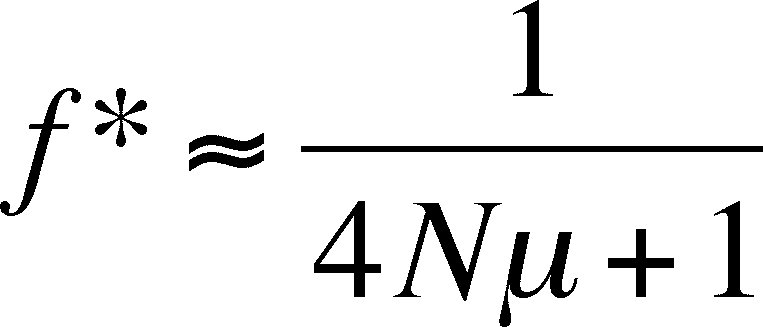
![]()
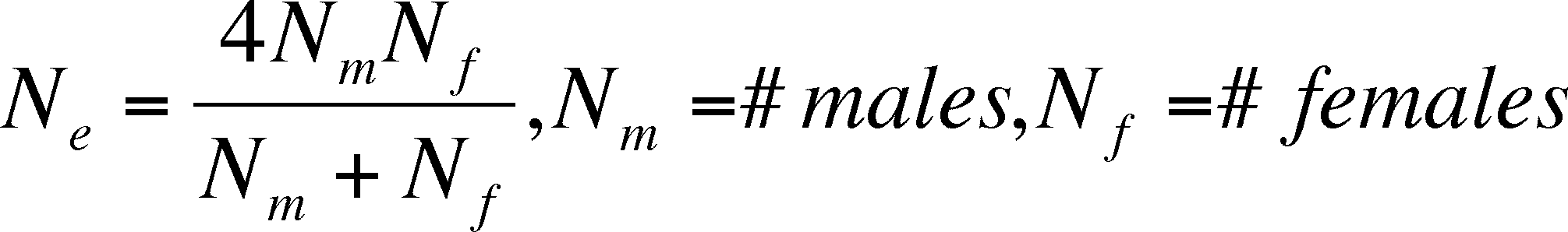
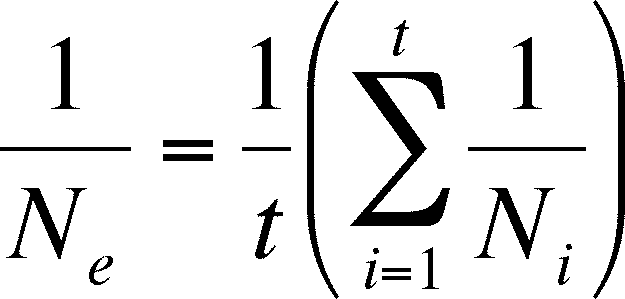
![]()